Description
INTRODUCTION: Parainfluenza virus 5 (PIV5), previously known as simian virus 5 (SV5), was first isolated in 1954 from primary monkey kidney cells, hence the original name SV5. Subsequent studies indicate that PIV5 is not a simian virus, and the virus was renamed PIV5 by International Committee on Taxonomy of Viruses in 2009. PIV5 has reportedly been isolated from a wide range of hosts including humans, dogs, pigs, cats, hamsters, and guinea pigs. PIV5 has been isolated on numerous occasions from human tissues and neutralizing antibody against PIV5 was detectable in symptomatic and asymptomatic humans. However, its association with human disease remains controversial. In addition, previous studies have not documented illness in infected animals, except kennel cough in dogs. PIV5 infects a large range of cell types including established cell lines as well as primary cells. Yet, PIV5 has very little cytopathic effect (CPE) on most infected cells. PIV5 is an enveloped non-segmented negative strand RNA virus and is a prototypical member of the Rubulavirus genus of the subfamily Paramyxovirinae of the family Paramyxoviridae. The PIV5 genome contains seven genes that encode eight known viral proteins: the nucleocapsid protein (N), the V protein (V) and phosphoprotein (P), the matrix protein (M), the fusion protein (F), the small hydrophobic integral membrane protein (SH), the hemagglutinin-nauraminidase (HN), and the large RNA-dependent RNA polymerase protein (L). The N, P, V, and L proteins are associated with viral genome to form the nucleocapsid core, the M protein resides between the core and viral envelope, and the SH, F, and HN proteins are integral membrane proteins, the latter two are involved in virus attachment and viral entry into cells. PIV5 is used as a prototype in the study of paramyxoviruses, and more recently, tested as viral vector candidate for vaccine development.
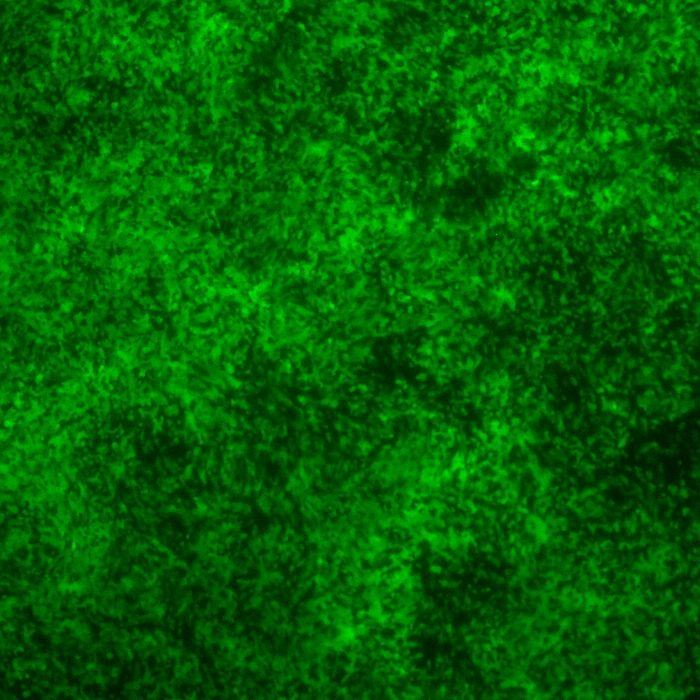
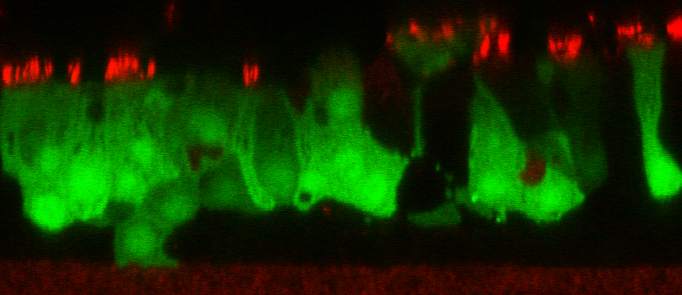
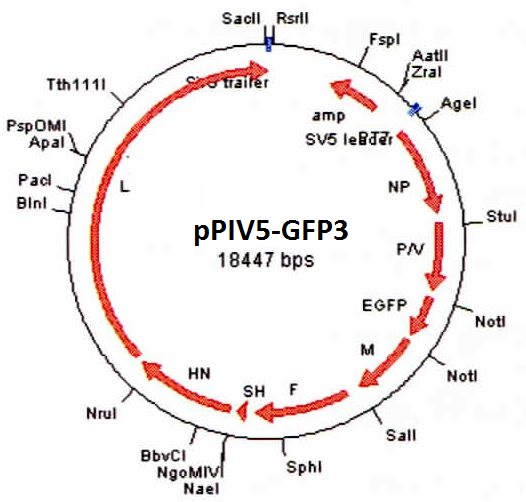
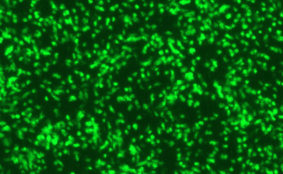
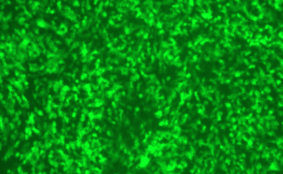
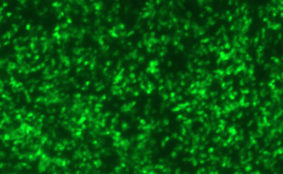
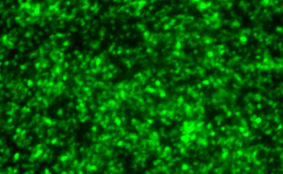
DESCRIPTION: A complete cDNA clone of the PIV5 genome of W3A strain was constructed such that an anti-genome RNA could be transcribed by T7 RNA polymerase and the correct 3’ end generated by cleavage using hepatitis delta virus ribozyme. To insert green fluorescent protein (GFP) gene into the PIV5 genome between V/P and M genes, the entire intergenic region between V/P and M was duplicated and the GFP gene (EGFP, Clontech) was inserted such that transcription of GFP is controlled by the first copy of V/P-M gene end and gene start sequences. The resulting full length pPIV5-GFP3 plasmid was designed so that the genome of the recovered virus obeys the rule of six. The pPIV5-GFP3 plasmid was transfected into BSR-T7/5 cells that stably expresses T7 polymerase, together with three support plasmids that expressed the viral replication proteins, N, P, and L, also under the control of the T7 polymerase promoter. Rescued virus was further propagated in LLC-MK2 cells. PIV5-GFP3 virus was found to replicate to similar titers as the wild-type virus in cell cultures.
| Parental Strain: | W3A strain |
| Construction: | GFP gene was inserted between V/P and M genes as the 3rd gene. |
| Passage History: | The isolate was propagated in LLC-MK2 cells. |
| Infectivity: | Titer > 8.0 log 10 TCID50 per mL. |
| Volume/Storage: | 2 x 1.2 mL per cryovial. Store at –80ºC. |
| Quality Testing: | No bacteria, fungus, or mycoplasma detected. Endotoxin <10 EU/mL. |
| Availability: | Bulk quantity and custom orders are available. Contact info@viratree.com. |
Product Sheet: Product_P523
Certificate of Analysis: CoA_Lot1551
Material Safety Data Sheet (MSDS): MSDS_PIV